Unusually High Read Coverage in a Genomic Region
Abstruse
The rapid development of next generation sequencing (NGS) applied science provides a new chance to extend the scale and resolution of genomic research. How to efficiently map millions of short reads to the reference genome and how to brand accurate SNP calls are ii major challenges in taking full reward of NGS. In this article, we reviewed the current software tools for mapping and SNP calling and evaluated their performance on samples from The Cancer Genome Atlas (TCGA) project. We found that BWA and Bowtie are better than the other alignment tools in comprehensive performance for Illumina platform, while NovoalignCS showed the best overall performance for SOLiD. Furthermore, we showed that next-generation sequencing platform has significantly lower coverage and poorer SNP-calling performance in the CpG islands, promoter and 5′-UTR regions of the genome. NGS experiments targeting for these regions should have higher sequencing depth than the normal genomic region.
Introduction
The advent of Next Generation Sequencing (NGS) engineering has significantly advanced the sequence-based genomic research and its downstream applications1 which include, just not limit to, metagenomics, epigenetics, gene expression, RNA splicing and RNA-seq and ChIP-seq2,3. In the by three decades, the Sanger method4 has been applied in many meaning big-scale sequencing projects and is considered as a 'gilt standard' considering of its appropriate read length and high accuracy5. Then far, three NGS platforms, the Roche/454 GS FLX, the Illumina/Solexa Genome Analyzer and the Applied Biosystems SOLiD System, have attained world-wide popularity. NGS focuses on generating three to four orders of magnitude more sequences but with considerably less cost in comparison with the Sanger method on the ABI 3730xL platform (hereafter referred to as ABI Sanger)5,vi,7. Despite the recent advances of NGS technologies, information technology is not clear whether the sequencing coverage by the NGS is the same across unlike regions of the genome.
Later the curt reads are generated, the first footstep is to align them to the reference genome. To discover tumor genetic information through resequencing unlike command/case samples, the mapping procedure must be able to efficiently align millions of sequences generated. Alignment algorithms should be robust enough to sequencing errors, but exist able to notice true genomic polymorphisms2. To take total advantage of NGS, more and more efficient algorithms are designed to overcome the limitation of read length and not-uniform error score in NGS data.
Because of the tremendous volume of reads and the huge size of the whole reference genome, alignment speed and retention usage are the two bottlenecks in mapping NGS reads. Traditional algorithms, such as Smash8 and BLAT9, can perform the NGS alignment more precisely, but they usually take a few days even on computer grids, non to mention personal computers. The fourth dimension and price are ordinarily unaffordable for most biologists. Some other challenge is how to pick truthful hit from multiple hits. Generally many aligners will report all possible locations with the appropriate tags or selection a location heuristically. If the multiple hits cannot exist ranked for certain standard, it will brand the comparison between read and reference unreliable. Furthermore, since the sequencing genome is usually different from the reference genome, alignment algorithms should be robust enough to sequencing errors, but do not miss true genomic polymorphismstwo. To handle these challenges, a lot of short-read alignment programs have been developed in recent years. A cursory review of the popular programs is provided in supplementary materials and all of them are free for academic and non-commercial employ.
Based on the core alignment techniques used, the programs tin can be classified into three categoriesten,xi. The first category uses hashing tables and it can exist further divided into two sub categories, either hashing the reads then using the reference genome to scan the hash tabular array, such every bit RMAP12,13, MAQxiv, ZOOM15, SeqMap16, SHRiMP17 (for the updated version two, it hashes the genomeeighteen) and RazerSnineteen, or hashing the reference genome then using the set of input reads to scan the hash tabular array, such as MOM20, Novoalign, MOSAIK and BFAST21. ('Hash table' refers to a common data structure that is able to alphabetize complex and non-sequential information in a way that facilitates rapid searching.)
The second category of programs, such as Bowtie22 (which does not support gaps yet), BWA11,23 and SOAPv224, are based on the Burrows–Wheeler Transform (BWT)25. They can efficiently align short sequencing reads confronting a big reference sequence, allowing mismatches and gaps. These methods typically utilize the FM index data construction, proposed by Ferragina and Manzini, who introduced the concept that a suffix assortment is much more than efficient if it is created from the BWT sequence, rather than from the original sequence26. The FM index retains the suffix array'south ability for quick pattern search and is generally smaller than the input genome size27.
The third category is implemented past merge-sorting the reference subsequences and read sequences. The representative one in this category is Slider28, which is focused on the Illumina platform information. The characteristics of the chosen software and their output formats are summarized in Table i. Since the offset ii categories are predominately used, we take assessed the functioning of the representative software in the 2 categories.
Furthermore, authentic alignment is non sufficient to meet the needs of further scientific discovery for nearly resequencing projects. For example, the 1000 Genomes Project aims at sequencing more 1000 homo genomes to characterize the pattern of genetic variants (mutual and rare) (http://world wide web.1000genomes.org/). TCGA (http://cancergenome.nih.gov/) has been sequencing a large number of cancer and normal samples for dissimilar individuals, targeting at the genetic variations of tumor. To this end, the whole analysis pipeline should also include detecting genomic variations including unmarried nucleotide polymorphism (SNP), copy number variations (CNV), inversions and other rearrangements29. Although NGS provides a sequencing error score, it is difficult to distinguish true genetic variation from the sequencing error or mapping error30.
Currently, there are several methods available for calling SNPs from NGS data, including Pyrobayes31, PolyBayes32, MAQ14, SOAPsnp33, Varscan34, SNVMix35,36, SeqEM37 and Atlas-SNP229. Pyrobayes and PolyBayes recalibrate base calling from raw data and so implement a Bayesian approach that incorporates prior information with population mutation rates to detect SNP. MAQ derives genotype calls from a Bayesian statistical model that incorporates the mapping qualities. It measures the conviction that a read actually comes from the position it aligns to, error probabilities from the raw sequence quality scores, sampling of the two haplotypes and an empirical model for correlated errors at a site. SOAPsnp is besides based on the Bayes' theorem. Information technology first recalibrates the sequencing quality score to calculate the likelihood of genotype for each position with existing conversion matrix and then combines the prior probability for each genotype to infer the true genotype33. Varscan uses parameters such equally the overall coverage, the number of supporting reads, average base quality and the number of strands observed for each allele to predict genotypes34. SNVMix combines 3 Binomial mixture models to model allelic counts, nucleotide and mapping qualities of the reads and infers SNPs and model parameters with the expectation maximization (EM) algorithm36. In contrast, SeqEM estimates parameters in an adaptive mode. It uses the EM algorithm to numerically maximize the observed information likelihood with respect to genotype frequencies and the nucleotide-read fault rate based on the NGS data of multiple unrelated individuals37. Atlas-SNP is similar to SOAPsnp, but it infers systematic errors of base of operations substitutions on single reads by plumbing fixtures training datasets using a logistic regression model which identified read sequence-related covariates to the base of operations-quality score29.
We used 3 representative programs – MAQ(version 0.71), SOAPsnp(version 1.03) and SNVmix(version two-0.xi.eight-r4)- to telephone call SNP on the merged GBM alignment consequence in bam file format38 and assumed the genotype of each base to be in one of iii states: 'aa' as homozygous for the reference allele, 'ab' equally heterozygous and 'bb' every bit homozygous for the non-reference allele, with the latter ii genotypes constituting an SNP36. We compared the NGS analysis result with the SNPs detected by the Affymetrix genome-broad human SNP array 6.0, which was treated as the gold standard. According to the setting, a true positive SNP is a site whose genotype is chosen as 'ab' or 'bb' in array and a true negative SNP is 'aa'.
Results
Alignment performance
We evaluated the performance of sequence mapping software in aligning reads from the cancer genome atlas (TCGA) project39, including 2 × thirteen,326,195 paired-end reads (SRR018643) and 15,578,118 unmarried-cease reads (SRR018725) with length of 76bp each from the Glioblastoma multiforme (GBM) sample (SRS004141) sequenced on Illumina Genome Analyzer Two, two × 13,716,752 paired-stop reads (SRR018658) with length of 76bp each from blood derived normal sample (SRS004142) sequenced on Illumina Genome Analyzer 2 and one million single-end reads(SRR030482) with length of 50bp from the Serous Cystadenocarcinoma sample (SRS004260) sequenced on the AB SOLiD Organisation 3.0.(come across Method section for item)
To compare the aligner operation fairly, we adjusted the parameters of each exact lucifer programs to standardize the general filters: at most 5 mismatches in whole read or at most 2 mismatched in start 28 bases seed region (if supported. Consider average 10% fault base calling rate in 30bp 3′bp tail and basic 2-seed-mismatch maq-like policy). However, this filtering strategy does not fit well for Smith-Waterman based algorithms. Smith-Waterman based algorithms penalize all errors (insertion, deletion, mismatch, etc) quantitatively and summarize them into one mapping score for filtering. And if encountered with paired-end reads, the insert range should be set from 0bp to one,000bp. The setting for insert size is a very loose standard because the insert size for our pair-finish sample in the genomic library has an average length of 586bp with a standard deviation of 101bp. Default settings were used for the other parameters of each plan.
From previous experiments, input/output loads were not significant factors in total running time22, so merely CPU time was considered for assessment. We as well divided the CPU running time into ii parts, time for index and time for alignment. Considering the index is reusable, the expensive cost of indexing volition no longer exist in the application afterwards. We tested these software tools on a typical desktop workstation with a 2.66 GHz Intel cadre 2 processor Q9400 and 16GB of RAM and the system openSUSE 11.1. All programs run on a single thread.
The assessment results based on Illumina paired-finish data are summarized in Table 2. BWT based aligners, Bowtie and BWA demonstrated the best overall performance compared with other index based methods. Bowtie has counterbalanced alignment sensitivity, efficient CPU usage and memory consumption, finishing the job within ii and half hours with over 67.5% reads aligned. Compared with Bowtie, BWA needed 88% more time to do the alignment only with merely five% more than reads aligned (72.99%) in 2-seed-mismatch maq-like policy.
RMAP, ZOOM and Maq belong to the "hashing reads" category. Due to the huge volume of reads to deal with, their memory footprints are not flexible anymore, ranging from 8GB to 10GB, which may not exist viable for non-expert users. ZOOM beats the other "hashing reads" aligners significantly, using seven 60 minutes to complete alignment with 60% sensitivity. Maq reached a better sensitivity of 72.0%, just consumed 39 hr x min for alignment. Thus the alignment speed up by nigh 20 folds in bowtie than Maq for 76bp length reads and the Maq also got a slighter higher sensitivity than bowtie, which is consistent with the comparison in bowtie paper22. For the "hashing reference" aligners tested, as expected, they had the worst operation on the running time and retentiveness consumption when parallelized computing was not implemented; yet, due to the underlying Needleman-Wunsch (Novoalign) and Smith-Waterman (SHRiMP) exact search algorithms, they showed excellent sensitivity. SHRiMP had a sensitivity of 81.2%, which was almost twenty% college than Bowtie, only it took 100 times longer than Bowtie for alignment due to the thread and RAM limitation. We also evaluated the performance of the eight programs on Illumina single-end data from the same GBM sample ( Supplementary Table one ) and observed similar results. To validate that the sample phenotype does non affect the performance of the aligner, we tested one run from normal sample ( Supplementary Table 2 ), the relative rankings of retentiveness consumption and computing speed of each aligner are similar in both samples. Meanwhile, the differences on sensitivity in both samples besides have the like trends for all aligners, which should exist attributed to the heterogeneity of sample inner property and the variation in the sample distension stage.
For the SOLiD information, NovoalignCS showed the all-time overall performance. Different from the letter of the alphabet-space index, all aligners except ZOOM create color-space index for SOLiD data. On average they had a lower proportion of reads mapped compared with the Illumina data. The time for the extremely high sensitivity in SOLiD alignment of SHRiMP was more than 1000 times longer than Bowtie ( Supplementary Table 3 ).
The preferred output format for each plan is besides listed in Table 1. The Sequence Alignment/Map (SAM) format38 is designed to support both single and paired-end reads, including color space and base infinite reads from unlike platforms, which creates a well-divers interface between alignment and downstream assay.
Sequencing depth, CpG islands and genomic coverage
We investigated how many sequencing depths are required to cover the whole genome. Nosotros mapped 13 runs of the GBM sample SRS004141 in experiment SRX006310 to the reference man genome (UCSC genome browser man genome version hg18) with Bowtie. With the increase of sequencing depth, the percent of genome covered increases (Figure 1). At one fold sequencing coverage (ane fold coverage = human genome three.0 gigabases), only less than 50% of the genome was covered at least once and less than twenty% was covered at least twice. At x folds sequence coverage, virtually 90% of whole genome was covered and 83% was covered at least twice.
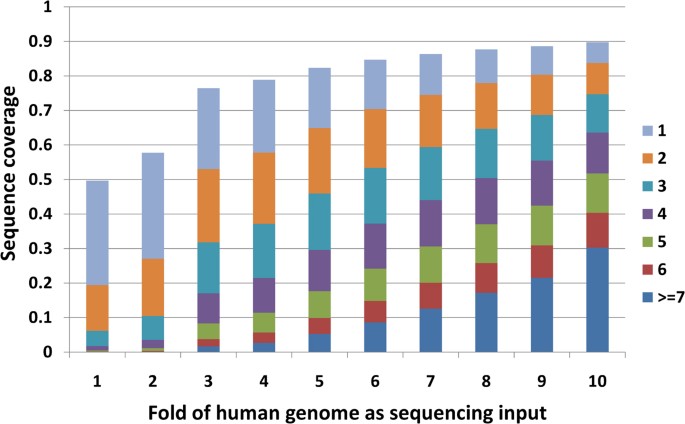
The relationship between sequence fold and genomic coverage.
Length of colour bar represents the per centum of bases with respective depth in the whole genome under corresponding volume of sequencing bases.
We next investigated whether unlike genomic regions have different coverage. Nosotros found CpG island regions accept meaning lower coverage than the whole genome and gene regions (both P values less than 2.2e-16). At ten folds coverage, only 50% of CpG islands were covered at least one time, compared to 90% for the whole genome (Figure two). Similarly, at one fold coverage, the numbers are 20% and fifty% respectively ( Supplementary Figure i ).
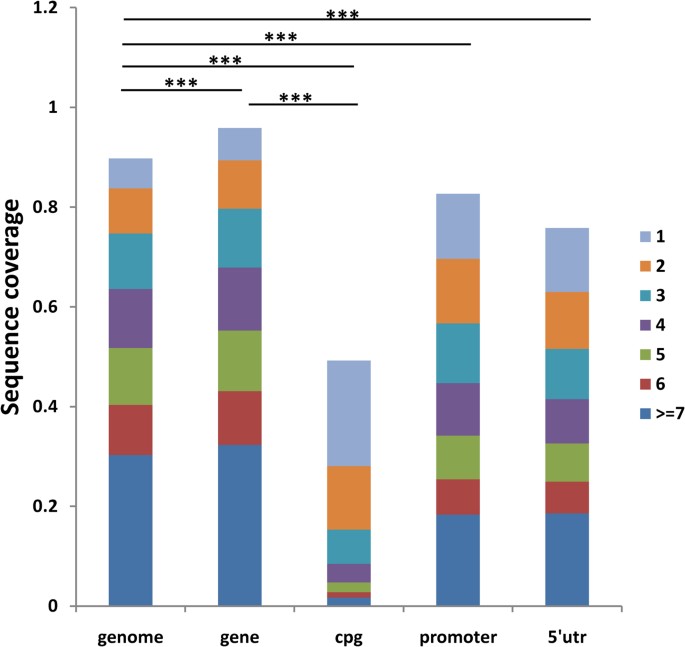
Coverage comparisons for different genetic regions at ten folds coverage.
P-value (all are less than 2.2e-16) for t-examination through bootstrap shows the significant poorer coverage of CpG-island region compared with genomic background or factor region. Meanwhile, the promoter and 5′UTR region are both significantly under-covered. (One star: p-value<0.05, 2 stars: p-value<0.01, three stars: p-value<0.001).
Since CpG islands are in 74% of upstream promoters and xl% of the downstream promoters of mammalian genesforty, nosotros hypothesized that, the promoter and five′UTR regions, which are important for regulatory roles of the genome, are also nether covered by the NGS technology. Indeed, in all three folds nosotros tested, promoter and v′UTR regions are significantly nether covered by next generation sequencing when compared with whole genome background ( Supplementary Effigy 2 ) (both P values less than 2.2e-16). At 10 folds coverage, merely 83% promoter and 76% v′UTR regions were covered at to the lowest degree one time, compared to xc% for whole genome. The numbers are 42%, forty% and l% respectively at one fold coverage.
Although factor region is well known to have a higher GC-content than the genome average, its coverage, unlike CpG-isle, is higher than the genome average. To further written report the human relationship between GC-content and sequence coverage, we randomly picked 10,000 windows with 1kb length each from human genome and computed their GC-content and sequence coverage at x fold coverage. Nosotros observed that sequence coverage increases with GC-content increment when GC-content is less than 40–45%, only decreases when GC-content is more than fifty–55%, with the summit at effectually 45% ( Supplementary Figure 3 ). This observation is consistent with previous discovery41. The CpG isle, promoter and 5′UTR regions take average GC-contents of 68.6%, 57.7% and 51.1%, which are higher than the peak at GC-content of 45%. This explains why all of them have sequence coverage lower than the genomic average. On the contrary, the gene region has average GC-content of 46%, which is at the peak. The figure explains why the coverage in that region is higher than whole genome average (with average GC-content of 41%).
Nosotros then investigated whether the repetitive element is also a factor causing low mappability in regulatory regions. For total 22571 promoter sequences, we ranked them by repetitive coverage (the portion of the sequence is covered past repetitive chemical element) then chose elevation 200 and lesser 200 sequences to compare their coverage pattern. Though the t-test showed significant difference between them (summit: 0.94±0.ten (mean ± std), lesser: 0.83±0.21, p-value: 7.33e-12), surprisingly, the sequences enriched for the repetitive element accept considerable higher coverage. We further ranked the promoters by GC-content then exercise the similar test. Nosotros institute that the acme 200 promoters have significantly lower coverage than the lesser 200 promoters (top: 0.ten±0.10, lesser: 0.92±0.xiii, p-value: 1.15e-222). The results indicated that the relatively college GC-content is the major cause of the lower coverage in regulatory regions.
SNP discovery performance
We chose loftier quality SNP probes as our test fix by removing the probes with a confidence score above 0.018. The exam set consisted of 583,891 probes, 98% (575,765/583,891) of which were covered by NGS when 10 folds coverage were used. The human relationship between NGS SNP coverage and genome fold coverage is shown in Figure iii. Under the default setting (SNVmix parameter was first set up as same equally that used in the original newspaper for the lobular chest tumor35, so trained itself by the model SNVmix2, which extended original Binomial mixture model SNVmix1. However, the genotype issue for cocky-training parameter showed like ROC performance, then we applied the first one), we obtained area under the ROC curve (AUC) (see Method). MAQ and SOAPsnp take similar results (AUC (MAQ) = 0.8872, AUC (SOAPsnp) = 0.8866) and both outperform SNVmix significantly (AUC (SNVmix) = 0.8394) (both P-value are < 2.2e-16).
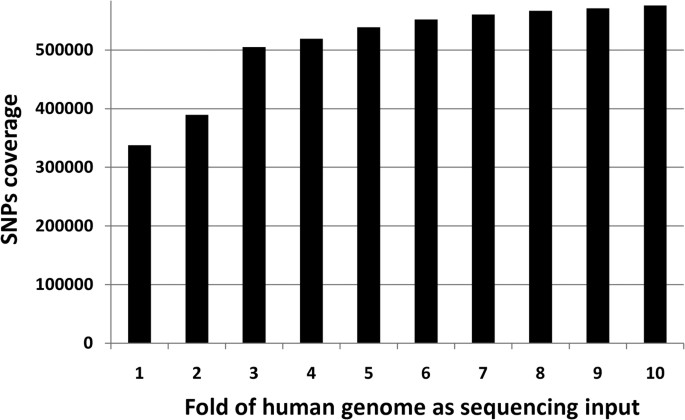
The relationship of the number of probes covered and genomic sequence fold (full 583891 SNP probes)
Nosotros also studied the SNP calling capability of the iii software tools on different depths (Figure 4). Due to the underlying post Bayesian concept, the accuracies of SOAPsnp and MAQ increase as the depth of the target bases increment. However, the SNVmix even demonstrated a worse operation under college coverage at 21–25 depths, which suggests its unstable performance without the cocky-training parameters. For depression-coverage SNPs, specially with the depth between one×–10×, the performances of MAQ still remain the best.
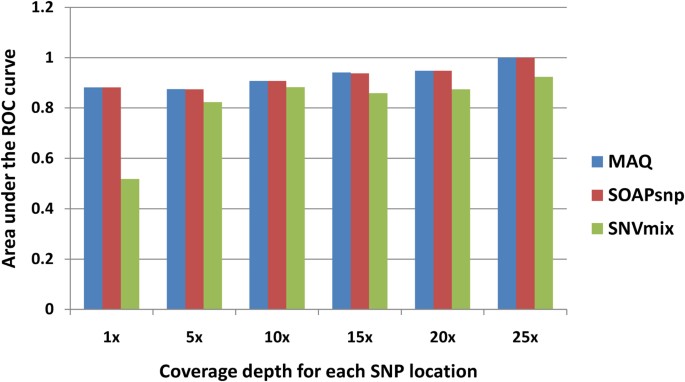
Comparison of SNP calling qualities (AUCs) of 3 software tools at different depths.
Alternatively, we calculated the overall genotype concordance which is divers in VariantEval module of the Genome Analysis Toolkit (GATK)42 to mensurate the understanding between SNPs called from NGS and assortment ( Supplementary Figure iv ). The concordance score was divers every bit (A+F+50)/ (A+B+C+E+F+G+I+J+50) ( Supplementary Table 4 ). The profile is similar to the AUC measurement, which shows that SNVmix is unstable in high depth situation. The low concordance score when coverage depth is under 20-fold suggests that there is still a big claiming to distinguish the heterozygote from the minor homozygote when sequence coverage is low.
Due to the poor sequence coverage in CpG-island, we tested the classifying performance for 711 SNP probes in array, which are located in CpG-island and covered by merged alignment files. The AUC for each method is, MAQ: 0.8429, SOAPsnp: 0.8379 and SNVmix: 0.5801. We farther tested performance for the promoter (3169 SNP probes) and 5′UTR region (1099 SNP probes) ( Supplementary Table 5 ). No matter which classifier was applied, performance in these regions was significantly junior to the genome background (P-value < 0.01) (Figure 5).
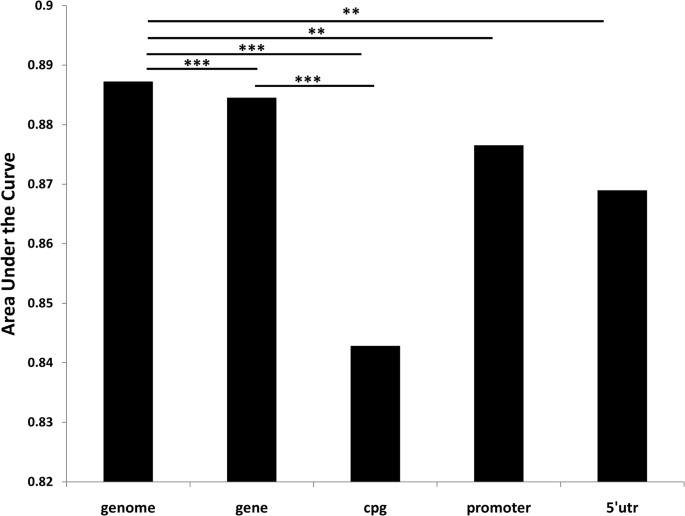
AUC (expanse nether the curve) comparison for different genetic regions.
CpG-island region has significantly poorer performance than genomic background (p-value = 0.000972) or gene region (p-value = 0.0003607). Promoter (p-value = 0.00873) and 5′UTR (p-value = 0.00946) region shows similar pattern. Gene-region too achieve a picayune fleck lower performance (p-value = 0.0004641).
Discussion
We have assessed 8 representative NGS mapping tools in aligning reads from the cancer genome atlas (TCGA) project. FM-alphabetize based aligners with BWT performed all-time in both paired-finish and single-end curt reads alignments. Evaluated on reads sequenced on the Illumina Genome Analyzer II, Bowtie demonstrated the all-time overall functioning. Bowtie has balanced alignment sensitivity, efficient CPU usage and retentivity consumption, finishing one run of sequences on the man genome within 2.5 hours with over 67.five% reads aligned. Meanwhile, nosotros should admit that a lot of aligners can run in muliti-thread manner in practical and hardware limitation may not be the barrier for normal groups. For example, the SHRiMP2 paper compare SHRIMP2, BFAST, BWA and Bowtie'southward performance on artificial data for different variation cases while utilizing an 8 cadre 3.0Ghz Intel Xeon machine with 16Gb RAM18. For that example, SHRiMP ii showed an adequate speed (20 folds slower than bowtie) and significantly higher precision and recall charge per unit. Thus if we primarily target the highly polymorphic reads and practise parallelization, these Smith-Waterman string matching algorithm based aligner should be our starting time choice in practice.
With bowtie every bit the aligner, 90% of the whole genome were at to the lowest degree once and 83% were covered at list twice when 10 folds (30 gigabases) input was given. Our results bear witness that three folds may be a minimum requirement for input raw data to reach more than than 50% of whole genome coverage.
In addition, we found that the CpG-island region shows a significantly poor coverage compared with the whole genome average. The promoter and five′UTR regions, which harbor regulatory elements and are closely associated with CpG islands40,43, are also significantly under-covered by NGS compared with the whole genome. Thus to discover above regions with target depth, we need to increase the number of runs. For case, to embrace 50% of genomic region at to the lowest degree one depth, nosotros need only one fold of the whole genome. However, to cover CpG island regions with the aforementioned criteria, we need x folds of the whole genome ( Supplementary Figure 2 ).
We as well evaluated the SNP calling capability of 3 software tools and found that MAQ performed the all-time. Nosotros constitute that like to mapping coverage, SNP calling performance also vary in different genomic regions. The CpG islands, promoter and 5′UTR regions have significantly lower SNP calling functioning than the genome and factor body regions. For the SNP analysis, 10 folds input is enough for the standard evaluation, though for the classic Bayesian method, the higher sequencing depth, the more accurate the SNP call will be. We only evaluated the software's capability on detecting known SNPs covered by array, merely not on novel ones. Several groups accept pioneered in this direction44, simply how to evaluate the accuracy is nonetheless to be solved in practice. Due to the limitation of SNP array on the number and distribution of the probes, NGS based GWAS will become a better resolution on the diseases related bio-markers.
In summary, we assessed major NGS assay tools for sequencing mapping and SNP calling and institute that Bowtie is the best tool for mapping and MAQ the exam tool for SNP calling. Furthermore, nosotros plant that CpG rich regions, such as promoter and 5′UTR, where regulatory elements are usually located, are poorly covered by the NGS platforms. This discovery raises the concerns for NGS engineering, particularly when regulatory elements are the focused report regions. NGS experiments for studying these regions should accept higher sequencing depth than the normal genomic region.
Methods
Reads extraction
The sequences all in fastq (csfastq for SOLiD) format were extracted from the database of genotype and phenotype (dbGap) in NCBI past sequence read archive (SRA) toolkit. They were then mapped to the homo reference genome [NCBI build 36.3] through assigned aligners. The real data was non filtered or modified (besides trimming) from what they originally appeared in SRA.
Coverage comparison for genomic regions
five′utr, iii′utr and gene regions were straight retrieved from refGene table in RefSeq genes track for hg18 through UCSC genome browser. Promoter regions were divers as starting at 5kb upstream of the transcriptional get-go site, ending at the terminate coordinate of the gene. CpG islands regions were retrieved from cpgIsland table in CpG Islands track for hg18 through UCSC genome browser. The repetitive elements were downloaded from the RepMask 3.2.7 track in UCSC genome browser. Genome background regions were simulated by randomly picking 10000 windows with 1kb length each from hg18 homo genome. Each original genomic region entry was in browser extensible data (BED) format. Then nosotros filtered the redundant entry and merged the overlapped entries together for each genomic feature. For each entry, we computed the coverage percent from the merged NGS alignment files. Then we figured out the boilerplate coverage for each genomic characteristic.
To validate the significance of difference between coverage of different genomic features, firstly we did 1000 times bootstrap to go 1000 sets of coverage entries of each genomic feature (each time with 80% volume of total entry number in the feature category). And so we did ii-sided t-exam for comparison between 2 features to go the P-value.
Performance test for the SNP-caller
For SOAPsnp and MAQ, we assigned the Phred-scaled likelihood that the genotype is identical to the reference, which is besides called 'SNP quality', equally predictor and assigned the 1 and 2 genotype in Affymetrix assortment as SNP example and 0 in genotype as SNP command for the response. Nosotros also did the 0 to 2 and ii to 0 conversion when the minor allele is the reference allele, before ROC display and AUC calculation. SNVmix outputs 3 possibilities, homozygous to reference, heterozygous genotype and homozygous to the non-reference, nosotros added the latter ii (AB and BB) together to go the 'SNP possibility' as predicator and as well assigned the 1 and 2 genotype in Affymetrix array as example and 0 in genotype as control for the response. To provide statistical significance for the comparison between dissimilar classifiers, firstly we constitute the genomic location which is both covered by SNP array and the NGS alignment method (total 583891 in Figure 3), then we did bootstrap 1000 times to go 1000 AUC values for each classifier (each fourth dimension with 80% volume), lastly we did 2 sided t-test to get the p-value. To compare the performance of classifier in dissimilar regions (the regions for each characteristic were defined as above), we do the like: firstly we found those coordinates which located in certain features and both covered by array and NGS alignment, then for each feature, we did bootstrap to get 1000 AUC values from the method, lastly nosotros did the same two sided t-test to compare different features to get the p-value.
References
-
Flicek, P. & Birney, E. Sense from sequence reads: methods for alignment and assembly (vol 6, pg S6, 2009). Nat Methods 7, 479–479 (2010).
-
Mardis, Due east. R. The impact of side by side-generation sequencing technology on genetics. Trends Genet 24, 133–141 (2008).
-
Mardis, East. R. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet nine, 387–402 (2008).
-
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with concatenation-terminating inhibitors. Proc Natl Acad Sci U S A 74, 5463–5467 (1977).
-
Bonetta, Fifty. Genome sequencing in the fast lane. Nat Methods three, 141–147 (2006).
-
von Bubnoff, A. Next-generation sequencing: the race is on. Jail cell 132, 721–723 (2008).
-
Schuster, S. C. Next-generation sequencing transforms today's biology. Nat Methods 5, sixteen–18 (2008).
-
Altschul, S. F., Gish, W., Miller, Westward., Myers, Due east. W. & Lipman, D. J. Basic Local Alignment Search Tool. J Mol Biol 215, 403–410 (1990).
-
Kent, W. J. BLAT–the Boom-similar alignment tool. Genome Res 12, 656–664 (2002).
-
Li, H. & Homer, N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform eleven, 473–483 (2010).
-
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
-
Smith, A. D., Xuan, Z. Y. & Zhang, G. Q. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics 9, 128 (2008).
-
Smith, A. D. et al. Updates to the RMAP short-read mapping software. Bioinformatics 25, 2841–2842 (2009).
-
Li, H., Ruan, J. & Durbin, R. Mapping brusque DNA sequencing reads and calling variants using mapping quality scores. Genome Research 18, 1851–1858 (2008).
-
Lin, H., Zhang, Z., Zhang, Chiliad. Q., Ma, B. & Li, M. ZOOM! Zillions of oligos mapped. Bioinformatics 24, 2431–2437 (2008).
-
Jiang, H. & Wong, W. H. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24, 2395–2396 (2008).
-
Rumble, Due south. Thousand. et al. SHRiMP: Accurate Mapping of Short Color-space Reads. Plos Comput Biol five, e1000386 (2009).
-
David, 1000., Dzamba, M., Lister, D., Ilie, L. & Brudno, M. SHRiMP2: Sensitive nevertheless Practical Short Read Mapping. Bioinformatics 27, 1011–1012 (2011).
-
Weese, D., Emde, A. K., Rausch, T., Doring, A. & Reinert, Yard. RazerS-fast read mapping with sensitivity control. Genome Research 19, 1646–1654 (2009).
-
Eaves, H. L. & Gao, Y. MOM: maximum oligonucleotide mapping. Bioinformatics 25, 969–970 (2009).
-
Homer, Northward., Merriman, B. & Nelson, Southward. F. BFAST: An Alignment Tool for Large Scale Genome Resequencing. Plos One 4, A95–A106 (2009).
-
Langmead, B., Trapnell, C., Pop, G. & Salzberg, Southward. L. Ultrafast and retentiveness-efficient alignment of short Dna sequences to the human genome. Genome Biology 10, R25 (2009).
-
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
-
Li, R. Q. et al. SOAP2: an improved ultrafast tool for curt read alignment. Bioinformatics 25, 1966–1967 (2009).
-
Basti, Yard. & Perrone, A. L. A fast hybrid block-sorting algorithm for the lossless interferometric data compression. P Soc Photo-Opt Ins 5103, 92-100228 (2003).
-
Ferragina, P. & Manzini, G. Opportunistic data structures with applications. Ann Ieee Symp Constitute, 390-398688 (2000).
-
Graf, S. et al. Optimized design and cess of whole genome tiling arrays. Bioinformatics 23, I195–I204 (2007).
-
Malhis, N., Butterfield, Y. South. North., Ester, Thousand. & Jones, S. J. 1000. Slider-maximum utilize of probability information for alignment of brusk sequence reads and SNP detection. Bioinformatics 25, 6–13 (2009).
-
Shen, Y. F. et al. A SNP discovery method to assess variant allele probability from side by side-generation resequencing data. Genome Enquiry 20, 273–280 (2010).
-
Cock, P. J. A., Fields, C. J., Goto, N., Heuer, Thousand. L. & Rice, P. M. The Sanger FASTQ file format for sequences with quality scores and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38, 1767–1771 (2010).
-
Quinlan, A. R., Stewart, D. A., Stromberg, G. P. & Marth, G. T. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat Methods five, 179–181 (2008).
-
Marth, G. T. et al. A general approach to unmarried-nucleotide polymorphism discovery. Nat Genet 23, 452–456 (1999).
-
Li, R. Q. et al. SNP detection for massively parallel whole-genome resequencing. Genome Research 19, 1124–1132 (2009).
-
Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
-
Shah, South. P. et al. Mutational evolution in a lobular chest tumour profiled at single nucleotide resolution. Nature 461, 809–U867 (2009).
-
Goya, R. et al. SNVMix: predicting single nucleotide variants from adjacent-generation sequencing of tumors. Bioinformatics 26, 730–736 (2010).
-
Martin, Due east. R. et al. SeqEM: An adaptive genotype-calling arroyo for next-generation sequencing studies. Bioinformatics 26, 2803–2810 (2010).
-
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
-
McLendon, R. et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
-
Wang, J. W., Ungar, Fifty. H., Tseng, H. & Hannenhalli, Due south. MetaProm: a neural network based meta-predictor for alternative man promoter prediction. BMC Genomics 8, 374 (2007).
-
Harismendy, O. et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biology 10, R32 (2009).
-
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing side by side-generation Deoxyribonucleic acid sequencing data. Genome Research twenty, 1297–1303 (2010).
-
Antequera, F. & Bird, A. Number of Cpg Islands and Genes in Human and Mouse. P Natl Acad Sci USA 90, 11995–11999 (1993).
-
Bansal, V. et al. Authentic detection and genotyping of SNPs utilizing population sequencing data. Genome Res twenty, 537–545 (2010).
Acknowledgements
Financial support was provided by Grants from the Research Grants Council (781511M, 778609M, N_HKU752/x) and Food and Wellness Bureau (10091262) of Hong Kong.
Writer information
Affiliations
Contributions
J.Due west. and West.W. designed studies, analyzed data and wrote the manuscript. West.West. performed experiments. Due west.Z. and T.W.L. provided guidance for the various functional areas.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This piece of work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
Reprints and Permissions
Almost this article
Cite this article
Wang, Due west., Wei, Z., Lam, TW. et al. Next generation sequencing has lower sequence coverage and poorer SNP-detection adequacy in the regulatory regions. Sci Rep 1, 55 (2011). https://doi.org/10.1038/srep00055
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/srep00055
Further reading
Comments
Past submitting a comment you lot concur to abide by our Terms and Community Guidelines. If you lot find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Source: https://www.nature.com/articles/srep00055
0 Response to "Unusually High Read Coverage in a Genomic Region"
Post a Comment